| Author | Affiliation |
|---|---|
| Sara W. Johnson, MD | Keck School of Medicine of the University of Southern California, Los Angeles, CA |
| Sean Henderson, MD | Keck School of Medicine of the University of Southern California, Los Angeles, CA |
ABSTRACT
Introduction:
Emergency department (ED) patients with venous thromboembolism (VTE) are eventually treated with a standard dose of warfarin despite the fact that a number of patients are known to be sensitive to warfarin and may experience supra-therapeutic INRs and adverse bleeding events. Pharmacogenetics is an emerging field of medical practice that seeks to improve drug safety and efficacy in an individual patient by tailoring treatment to the patient’s known genetic makeup. To identify patients with risk for warfarin sensitivity among an ED population with VTE and to assess if the warfarin sensitivity mutations were of significant enough prevalence to be of clinical significance in customizing treatment of VTE. We sought in a pilot study to identify if testing for common CYP2C9 and VKORC1 single nucleotide polymorphisms (SNPs) in patients who were likely to begin warfarin treatment was feasible in an ED setting.
Methods:
A prospective study that identified and enrolled patients presenting to our ED with high clinical suspicion of VTE. Those with high clinical suspicion of VTE were defined as those who had a Doppler ultrasound or computed tomography pulmonary angiography (CTPA) ordered by the primary emergency physician. Blood was taken and processed to ascertain the following SNPs: CYP2C9*2, CYP2C9*3, and VKORC1 3673.
Results:
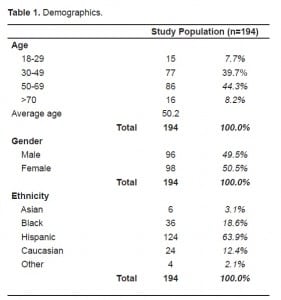
Of the 194 patients enrolled, 132 (68.0%) had at least one known warfarin sensitivity mutation and 114 (58.8%) had the most clinically significant VKORC1 3673 mutation.
Conclusion:
A majority of our patients had at least one mutation associated with the atypical metabolism of warfarin. Over half of our population had the most clinically significant VKORC1 3673 mutation. They would likely benefit from individualized warfarin dosing if ever needing anticoagulation. Our initial pilot study shows that allele frequencies of target warfarin sensitivity SNPs in our patient population are frequent enough to make initiation of personalized warfarin dosing feasible.
INTRODUCTION
Warfarin sodium is the most popular coumarin anticoagulant used worldwide. In 2004, 31 million dispensed outpatient prescriptions were written for warfarin in the United States (U.S.) alone. This number represents a 45% growth in the number of warfarin prescription from only six years prior and the number continues to rise.1 Warfarin unfortunately also boasts one of the highest risk profiles for any drug on the market. Its extensive use, along with its high rate of adverse events, adds a significant amount of morbidity and mortality in the U.S. annually. Despite its drawbacks, warfarin remains standard treatment for venous thromboembolism (VTE) because it is effective and inexpensive. The typical emergency physician’s use of warfarin is limited to initiating therapy for the occasional low-risk VTE patient who will be treated as an outpatient, as well as adjusting dosage in the setting of an abnormally high or low INR.
The significant amount of morbidity and mortality attributed to warfarin has made it a favorite drug of focus for the growing pharmacogenetics movement. Pharmacogenetics is an emerging field of medical practice that seeks to improve drug safety and efficacy in an individual patient by tailoring pharmacologic treatment to the patient’s known genetic makeup. Pharmacogenetics will undoubtedly produce many novel applications in emergency medicine and personalized warfarin dosing is likely to be among the first. Were emergency clinicians better able to predict a patient’s individual response to warfarin treatment when initiating or adjusting therapy, the risk and incidence of adverse effects should be diminished. This knowledge may also serve to increase the proportion of patients with newly diagnosed VTE who are safe to initiate oral anticoagulation at home with close outpatient follow up instead of inpatient warfarin initiation.
The pharmacogenetic focus for warfarin has primarily been on two enzymes involved in its mechanism of action and metabolism: the Vitamin K epoxide reductase complex subunit 1 (VKORC1) and the cytochrome P450 2C9 (CYP2C9) enzyme. We sought to identify in a pilot study if testing for common CYP2C9 and VKORC1 single nucleotide polymorphisms (SNPs) in patients who were likely to begin warfarin treatment was feasible in an emergency department (ED) setting. When researchers utilized VKORC1 mutations, CYP2C9*2, CYP2C9*3, and a number of demographics (age, body weight and concomitant medications) in a Swedish population, they were able to account for 50–60% of warfarin dose variability.2 By adding knowledge of an individual’s Protein C SNPs they were able to increase the dose variation prediction to 62%. Adding on a number of other additional SNPs of minor molecules involved in warfarin’s action only increased the prediction to 73%. This showed that while a better prediction of dose variation could be obtained by genotyping many additional SNPs, the majority of warfarin dose variation can be explained by simple patient demographics and the VKORC1, CYP2C9*2, and CYP2C9*3 mutations.3 As such, the VKORC1 3673 (1639G>A), CYP2C9*2 (430C>T), and CYP2C9*3 (1075A>C) SNPs served as the focus of our investigation. Little is known about the allele frequencies of CYP2C9*2, CYP2C9*3 and VKORC1 3673 in a Hispanic population; we also sought to investigate if the allele frequencies in our predominately Hispanic patient population were high enough to warrant testing for such mutations.
METHODS
This study was an initial pilot study of feasibility. As such we conducted a cross-sectional study with convenience sampling of an ED population with suspected VTE. We chose to study a patient population with suspected thromboembolic disease because we believed VTE diagnosed in the ED would be the most likely time an emergency physician would initiate warfarin therapy. As our selected mutations involve a patient’s susceptibility to warfarin sensitivity and not to VTE susceptibility, we did not expect to see any difference in the mutation rates between those patients eventually diagnosed as VTE positive and those VTE negative.
Our study design and protocol was approved by the University of Southern California Health Sciences Institutional Review Board. The authors have no conflicts of interest to report. Trained research assistants identified eligible patients in the LAC+USC Emergency Department and inpatient wards for enrollment. The enrollment time period was from April 2006 to July 2008. Our inclusion criteria were age ≥ 18, ability to give informed consent, in either English or Spanish, and having received an ultrasound of the lower extremities or computed tomography pulmonary angiogram (CTPA) ordered from the ED. Our only exclusion criterion was not satisfying all inclusion criteria.
Trained research assistant obtained informed consent for patients willing to participate, using either English or Spanish documents and consent forms. The study participants were asked a brief focused medical history and to self-identify their ethnicity as White, Hispanic, Black, Asian or other. Blood samples were obtained for each participant. DNA extraction was accomplished using the Qiagen 96 DNA Blood Biorobotic Kit. Genotyping was done using the Taqman 5′ Nuclease Assay looking for the following polymorphisms: CYP2C9*2 (430C>T) (rs1799853), CYP2C9*3 (1075A>C) (rs1057910), VKORC1 3673 (1639 G>A) (rs9923231). For each polymorphic site, two oligonucleotide fluorogenic minor groove binding probes were designed using Primer Express 2.0 Software (Applied Biosystems). We read and analyzed the fluorescence patterns of the PCR samples using an ABI 7900HT Sequence Detection System and Sequence Detection Software (Applied Biosystems). For reporting purposes, the CYP2C9*2 and *3 haplotypes are often combined in the literature and written as *1/*1 (for wild type), *1/*2, *1/*3, *2/*2, *2/*3, and *3/*3.
RESULTS
Of the 194 patients enrolled, 132 (68.0%) had at least one warfarin sensitivity mutation of either the CYP2C9 enzyme or VKORC1 receptor (Table 2). We had 20 patients (10.3%) with two mutations and two patients (1.0%) with all three mutations. For the most clinically significant VKORC1 3673 (1639G>A), we had 114 patients (58.8%) with mutations, 86 (44.3%) of whom were heterozygote variants (GA) and 28 (14.4%) of whom were homozygote variants (AA).
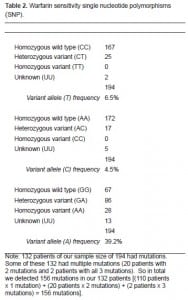
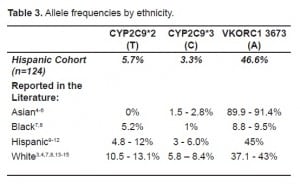
Our study also offered a unique opportunity to test the allele frequencies of a predominantly Hispanic cohort of patients. Approximately 64% (n=124) of our population self-identified as Hispanic. Calculated variant allele frequencies for our Hispanic cohort were: 5.73% for CYP2C9*2 T allele, 3.28% for CYP2C9*3 C allele, and 46.64% for VKORC1 3673 A allele (Table 3).
DISCUSSION
In 2002 and again in 2004–2005, warfarin and insulin were the two most commonly identified medications associated with adverse events presenting to U.S. EDs.16–18 From 1999–2003 there were an estimated 29,000 ED visits in the U.S. for warfarin bleeding complications alone. Throughout the 1990s and 2000s warfarin has continually been in the FDA’s Adverse Event Reporting System’s top 10 most reported drugs.1 The frequency of “major bleeding” (leading to death or requiring hospitalization or transfusion) adverse effects for patients on warfarin varies by study but has been reported to be anywhere from 0–16%.19–21 From a survey of U.S. death certificates, anticoagulants were the most frequently mentioned drugs causing “adverse events in therapeutic use” in 2003 and 2004.
The pharmacogenetic focus for warfarin on CYP2C9 and VKORC1 has been because these two enzymes are key to warfarin’s mechanism of action and metabolism. Normally, Vitamin K is recycled by the liver using the VKOR enzyme complex. This allows for the efficient use of Vitamin K and allows for sufficient stores of the reduced forms of the Vitamin. Reduced Vitamin K is the main cofactor used by gamma-glutamyl carboxylase (GGCX) to activate Vitamin K-dependent proteins: factors II, VII, IX, and X and proteins C and S. Warfarin acts by blocking the VKORC subunit 1 (VKORC1), thereby preventing the recycling of Vitamin K. This leads to a depletion of the body’s Vitamin K stores and decreased activation of Vitamin K-dependent clotting factors. Inactivated Vitamin-K dependent proteins are eventually secreted from the body.3 Warfarin sodium, as administered, generally consists of a racemic mixture of the R-enantiomer and S-enantiomer of the drug. The S-form is 3–5 times more active than the R-form.22 Once in the liver, S-warfarin is primarily metabolized by the cytochrome P450 2C9 (CYP2C9) enzyme.3 While other cytochrome P450enzymes metabolize R-warfarin, since it is the less active form, these enzymes are not as clinically significant to the pharmacokinetics of warfarin.
Numerous researchers have explained up to 50–60% of the variance in warfarin doses seen among patients, using the common VKORC1 and CYP2C9 SNPs and known environmental factors.2,14,15,23–28 Mutations in the VKORC1 gene alone are reported to account for approximately 25–30% of warfarin dose variance.2,15,29 CYP2C9*3(1075A>C) mutation is a SNP substitution of a normal adenine with a cytosine at nucleotide position 1075 of the gene. This mutation severely impairs the CYP2C9 enzyme from hydroxylating s-warfarin, leading to a 27-fold decrease in enzyme efficiency and 71–97% reduced warfarin clearance.4,30 The CYP2C9*3 mutation alone contributes approximately 10–12% to warfarin dose variance.3,15,29,30 The CYP2C9*2(430C>T) mutation is a SNP substitution of a normal cytosine for thymine at nucleotide position 430 of the gene. This SNP also results in reduced warfarin metabolism, but only an approximate six-fold reduction in enzyme efficiency and 29–58% reduced warfarin clearance.4,31,32
The specific CYP2C9*2 SNP contributes only 2.5% to warfarin dose variance.2 However, many other CYP2C9 SNPs are in strong linkage disequilibrium with CYP2C9*2, and testing for the *2 SNP likely continues in the literature because it represents the best way to test for all the other SNPs in linkage disequilibrium with it. When researchers tested 55 other CYP2C SNPs, only one added more to dose variation prediction than simply using CYP2C9*2 and CYP2C9*3 alone.3 So, testing of the CYP2C9*2 and *3 SNPs remains a good way to approximate an individual’s CYP2C9 phenotype.
There are an increasingly large number of newly discovered mutations of the VKORC1 receptor. One of the most commonly studied mutations is the VKORC1 3673(1639G>A). This SNP substitutes a normal guanine with an adenine at nucleotide position 1639 of the gene. Because this SNP has been found to be in strong linkage disequilibrium (r2>0.9) to a number of other VKORC1 SNPs significantly associated with warfarin dosing (381, 6484, 6853, and 7566) it is a good candidate for testing all of these mutations.29 It is believed that individuals with mutation VKORC1 enzymes have lower baseline enzyme activity and therefore lower baseline levels of active Vitamin K-dependent clotting factors.33 These individuals would already be predisposed to spontaneous bleeding, and the addition of an “average” warfarin dose could push these individuals to supra-therapeutic INR levels. In a study by Sconce et al14, those with the homozygous variant mutations for VKORC1 3673 required only 2.23 mg/day warfarin versus the heterozygote variants averaging 3.83 mg/day and homozygous wild type averaging 4.53 mg/day.
It is interesting to note that at least 68% of our patient population at risk for warfarin adverse effects had at least one warfarin sensitivity mutation of either CYP2C9 or VKORC1. Further, 58.8% had the VKORC1 mutation, which is known to be the most clinically significant contribution to warfarin sensitivity. This means that at least this 58% of our population would likely have benefited from a decreased initial warfarin dose than standard algorithms would have prescribed. Although our study specifically looked at VTE patients, we would expect to find similar mutations rates for all patients needing warfarin anticoagulation (atrial fibrillation, prosthetic heart values, pre-op patients, etc.). However, mutation rates for these specific populations need further testing before such assumptions can be verified. Our pilot study also showed that testing for warfarin sensitivity SNPs in the ED for patients prior to starting the medication may be feasible once more rapid testing is widely available. The allele frequencies of the VKORC1, CYP2C9*2, and CYP2C9*3 in our population were high enough to warrant testing for these mutations.
Our study also helped add to the knowledge of VKORC1 3673, CYP2C9*2, and CYP2C9*3 allele frequencies in a Hispanic-American population. Review of the literature revealed only three prior studies looking at Hispanic CYP2C9 allele frequencies and one additional study to include VKORC1 allele frequencies in a Hispanic population.9–12 Two studies looked at South American populations and found allele frequencies of 4.8% for CYP2C9*2 and 3.0% for CYP2C9*3 in 778 Bolivians and VKORC1 3673 frequencies of 45% for 191 genetically isolated Columbians.9,11 Another study looked at 434 Hispanic-Americans and found allele frequencies of 12.0% for CYP2C9*2 and 3.4% for CYP2C9*3.12 Finally, one study specifically looked at the allele frequencies differences between Mexican-Americans and Spaniards and found that 196 Mexican-Americans had allele frequencies of 8.0% for CYP2C9*2 and 6.0% for CYP2C9*3; these frequencies were statistically different from the 16.0% for CYP2C9*2 and 10.0% for CYP2C9*3 for Spaniards and pointed to the need for optimizing dosing based on Hispanic-American allele frequency data instead of applying European or Caucasian allele frequencies to Hispanic populations.10 The allele frequencies for our Hispanic population of 5.73% for CYP2C9*2, 3.28% for CYP2C9*3, and 46.64% for VKORC1 3673 were similar to these other studies. From what is known of these warfarin SNPs in the Hispanic population, the allele frequencies most closely align with the Caucasian population. However, it should be noted that the Hispanic population tends to have slightly lower CYP2C9*2 and higher VKORC1 3673 frequencies than the Caucasian population (Table 3). The wide inter-ethnic variability of allele frequencies for CYP2C9*2, CYP2C9*3, and VKORC1 3673 has previously been shown to explain much of the inter-ethnic variability in warfarin dosing.8,6,12,26,28
Finally, a recent multi-national NIH funded study published in the New England Journal of Medicine34 has used data on over 4,000 patients to establish a genetics-based dosing algorithm for warfarin. This algorithm uses basic patient characteristics, such as age, gender, and ethnicity, along with the patient’s known CYP2C9 and VKORC1 SNPs to more accurately predict a patient’s warfarin maintenance dose than standard dosing algorithms. While the algorithm has been validated, it is not yet known if using this new pharmacogenetic-based algorithm will decrease the cost of warfarin treatment and dosing titration as well as the drug’s adverse effects.
A number of small studies have indicated a decreased incidence of warfarin adverse events when genetics-based dosing algorithms are used in initiating warfarin therapy versus standard dosing algorithms.35,36 However, a few other studies have failed to show a statistical difference in adverse events and outcomes between genetic-based and standard algorithms.37 Larger randomized controlled studies will be needed to determine the superior algorithm and the cost-benefit analysis of using pharmacogenetic-based warfarin dosing. Also, while a few of the small randomized controlled studies that have been completed have boasted a quick turn-around time of genotyping in 1–4 hours, these studies still used research laboratories for testing.24,35–37 Clinical laboratory and point of care testing for both the CYP2C9 and VKORC1 SNPs are being developed and will likely reach mainstream markets in a year or two. Once this occurs we are likely to see increased use of pharmacogenetic-based warfarin dosing.
CONCLUSION
Knowledge of a patient’s warfarin sensitivity SNPs is likely to benefit emergency medicine by decreasing the number of inpatient admissions for warfarin titration and decreasing ED visits for warfarin adverse events. Practitioners can well imagine a clinical scenario in which a patient is low risk for VTE complications but high risk for anticoagulation complications, for example the elderly. Knowledge of warfarin sensitivity SNPs would allow the emergency physician to tailor the warfarin dose specifically for the patient, thereby decreasing the risk of elevated INR and bleeding. The patient might then be safely discharged home with outpatient follow up instead of admission. As technology for genetic testing improves, a patient’s warfarin susceptibility SNPs might become part of an emergency physician’s evaluation and treatment of VTE. Our pilot study shows that allele frequencies of target warfarin sensitivity SNPs are frequent enough to make personalized warfarin dosing in the ED feasible.
Footnotes
Supervising Section Editor: H. Bryant Nguyen MD, MS
Submission history: Submitted March 25 2010; Revision received November 3, 2010; Accepted November 10, 2010
Reprints available through open access at http://escholarship.org/uc/uciem_westjem
Address for Correspondence: Sean O. Henderson, MD, Department of Emergency Medicine, LAC + USC Medical Center, 1200 N. State Street Room 1011, Los Angeles, CA 90033
Email: sohender@usc.edu
Conflicts of Interest: By the WestJEM article submission agreement, all authors are required to disclose all affiliations, funding sources, and financial or management relationships that could be perceived as potential sources of bias. The authors disclosed none.
REFERENCES
1. Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. Arch Intern Med. 2007;167(13):1414–9. [PubMed]
2. Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–70. [PubMed]
3. Wadelius M, Chen LY, Eriksson N, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. [PMC free article] [PubMed]
4. Obayashi K, Nakamura K, Kawana J, et al. VKORC1 gene variations are the major contributors of variation in warfarin dose in Japanese patients. Clinical Pharmacology & Therapeutics.2006;80(2):169–78. [PubMed]
5. Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;2:251–63. [PubMed]
6. Yuan H, Chen J, Lee MM, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet.2005;14(13):1745–51. [PubMed]
7. Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–31. [PMC free article] [PubMed]
8. Wang D, Chen H, Momary K, et al. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood.2008;112(4):1013–21. [PMC free article] [PubMed]
9. Bravo-Villa HV, Yamamoto K, Nakamura K, et al. Genetic polymorphism of CYP2C9 and CYP2C19 in a Bolivian population: an investigative and comparative study. Eur J Clin Pharmacol.2005;61:179–84. [PubMed]
10. Llerna A, Dorado P, O’Kirwan F, et al. Lower frequency of CYP2C9*2 in Mexican-Americans compared to Spaniards. Pharmacogenomics J. 2004;4:403–6. [PubMed]
11. Palacio L, Falla D, Tobon I, et al. Pharmacogenetic impact of VKORC1 and CYP2C9 allelic variants on warfarin dose requirements in a Hispanic population isolate. Clin Appl Thromb Hemost.2009;16(1):83–90. [PubMed]
12. Xie HG, Prasad HC, Kim RB, et al. CYP2C9 allelic variants: ethnic distribution and functional significance. Advanced Drug Delivery Reviews. 2002;54:1257–70. [PubMed]
13. Higashi MK, Veenstra DL, Kondo LM. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. [PubMed]
14. Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen.Blood. 2005;106:2329–33. [PubMed]
15. Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113(4):784–92. [PMC free article] [PubMed]
16. Budnitz DS, Pollock DA, Mendelsohn AB, et al. Emergency department visits for outpatient adverse drug events: demonstration for a national surveillance system. Ann Emerg Med.2005;45(2):197–206. [PubMed]
17. Budnitz DS, Pollock DA, Weidenbach KN, et al. National surveillance of emergency department visits for outpatient adverse drug events. JAMA. 2006;296(15):1858–66. [PubMed]
18. Hafner JW, Belknap SM, Squillante MD, et al. Adverse drug events in emergency department patients. Ann Emerg Med. 2002;39(3):258–66. [PubMed]
19. Da Silva MS, Sobel M. Anticoagulants: to bleed or not to bleed, that is the question. Semin Vasc Surg. 2002;15(4):256–67. [PubMed]
20. Linkins LA, Choi PT, Douketis JD. Clinical impact of bleeding in patients taking oral anticoagulant therapy for venous thromboembolism: a metaanalysis. Ann Intern Med. 2003;139:893–900.[PubMed]
21. Schulman S, Beyth RJ, Kearon C, et al. Hemorrhagic complications of anticoagulant and thrombolytic treatment: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition). Chest. 2008;133(6 Suppl):257S–98S. [PubMed]
22. Rettie AE, Korzekwa KR, Kunze KL, et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5:54–9. [PubMed]
23. Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P45 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. [PubMed]
24. Huang SW, Chen HS, Wang XQ, et al. Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patients.Pharmacogenetics and Genomics. 2009;19:226–34. [PubMed]
25. Lee SC, Ng SS, Oldenburg J, et al. Interethnic variability of warfarin maintenance requirement is explained by VKORC1 genotype in an Asian population. Clin Pharmacol Ther. 2006;79:197–205.[PubMed]
26. Takahashi H, Wilkinson GR, Nutescu EA, et al. Different contributions of polymorphisms in VKORC1 and CYP2C9 to intra- and inter-population differences in maintenance dose of warfarin in Japanese, Caucasians and African-Americans. Pharmacogenet Genomics. 2006;16:101–10.[PubMed]
27. Vecsler M, Loebstein R, Almog S, et al. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin.Thromb Haemost. 2006;95:205–11. [PubMed]
28. Veenstra DL, You JH, Rieder MJ, et al. Association of Vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics. 2005;15:687–91. [PubMed]
29. Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–93. [PubMed]
30. Haining RL, Hunter AP, Veronese ME, et al. Allelic variants of human cytochrome P450 2C9: baculovirus-mediated expression, purification, structural characterization, substrate stereoselectivity, and prochiral selectivity of the wild-type and I359L mutant forms. Arch Biochem Biophys. 1996;333:447–58. [PubMed]
31. Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH:cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7:203–10. [PubMed]
32. Rettie AE, Wienkers LC, Gonzalez FJ, et al. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. [PubMed]
33. Rost S, Fregin A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–41. [PubMed]
34. The International Warfarin Pharmacogenetics Consortium Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. [PMC free article] [PubMed]
35. Hillman MS, Wilke RA, Yale SH, et al. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clinical Medicine & Research.2005;3:137–45. [PMC free article] [PubMed]
36. Lenzini PA, Grice GR, Milligan PE, et al. Laboratory and clinical outcomes of pharmacogenetic vs. clinical protocols for warfarin initiation in orthopedic patients. Journal of Thrombosis and Haemostasis. 2008;6:1655–62. [PMC free article] [PubMed]
37. Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–70.[PubMed]